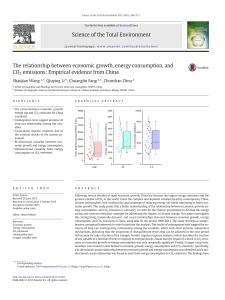
September 2016. Horticultural Plant Journal, 2 (5): 279–283. Horticultural Plant Journal Available online at www.sciencedirect.com The journal’s homepage: http://www.journals.elsevier.com/horticultural-plant-journal Karyotype Analysis of Gazania rigens Varieties ZENG Jiashi, WANG Dongxu, WU Yangqing, GUO Xiaoyu, ZHANG Yazhou, and LU Xiaoping * Gold Mantis School of Architecture, Soochow University, Suzhou, Jiangsu 215123, China Received 23 May 2016; Received in revised form 2 July 2016; Accepted 2 September 2016 Available online 25 November 2016 Abstract For studying species origin, systematic evolution and phylogenetic relationship of Gazania rigens, four different G. rigens varieties, with dif- ferent flower colors, were subjected to chromosome karyotype analysis. The somatic chromosome number in three varieties ‘Hongwen’, ‘Xingbai’ and ‘Richu’ was 2n = 10, while in ‘Zhongguo Xunzhangju’ it was 2n = 20. We speculate that the cardinal number of chromosomes in G. rigens plants is x = 5, in which case ‘Zhongguo Xunzhangju’ is a tetraploid. The karyotype formulae of ‘Hongwen’, ‘Xingbai’ and ‘Richu’ were 2n = 8m + 2sm, 2n = 8m + 2sm and 2n = 10m respectively. The karyotype formula of ‘Zhongguo Xunzhangju’ was 2n = 18m + 2sm. The asymmetrical karyo- type coefficients of the four G. rigens varieties ranged from 53.80% to 58.84%. Only ‘Richu’ had a ‘1A’ karyotype, while the others were relatively symmetric ‘2A’. Karyotype analysis indicates that the three introduced varieties have a close genetic relationship. Keywords: Gazania rigens; chromosome; karyotype analysis ). In the present research, we analyzed chromosome karyotypes four 1. Introduction Gazania varieties and found that the chromosome number in three Gazania rigens ( L.) Gaertn. is a member of the introduced varieties was 2n = 10, and for the variety ‘Zhongguo Asteraceae. It is a herbaceous perennial named for a flower shape Xunzhangju’, the number was 2n = 20. We propose that the chroresem- bling a medal. G. rigens is native to South Africa and mosome cardinal number in Gazania i s x = 5, and that ‘Zhongguo grows best in warm, sunny locations. Two types, tufted and Xunzhangju’ is a tetraploid. decum- bent, were introduced into China as useful groundcovers (Xie et al., 2013). They are evergreen with colorful flowers with a 2. Materials and methods long flower life and flowering season, and have a strong resistance v to drought, heat, poor soils and moderately cold temperatures. G. We used the methods of Li et al. (2008) and Zhu et al. rigens i s strongly agamogenic, making it easy to propagate by cuttings, plant division, and tissue culture, and it rapidly colonizes (2011). Root samples were taken when the new root length of roadbed landscapes. Li (2011) and Wang (2012) discussed cuttings from the Gazania varieties reached 1–3 cm. Roots were potential landscape applications. Wang (2013) and Zhou (2014) pre- treated for 20–24 h in ice water, then transferred into noted plant, leaf, flower shape, and flower color variability of G. Carnoy’s fluid (ethyl alcohol:glacial acetic acid = 3:1), immobilized for 20–24 h at 4 °C, washed three times with 90% rigens. alcohol, then trans- ferred into 70% alcohol, at 4 °C. Root tips The cytology of G. rigens is poorly known. Chen et al. were sliced and subjected to acidolysis by immersing in 1 mol·L−1 (2003) reported that Gazania w as diploid (2n = 20), and its hydrochloric chromo- some cardinal number was x = 10, but Barkley (2006) suggested that the chromosome cardinal number was x = 9 (2n = * Corresponding author. E-mail address: [email protected] Peer review under responsibility of Chinese Society for Horticultural Science (CSHS) and Institute of Vegetables and Flowers (IVF), Chinese Academy of Ag- ricultural Sciences (CAAS) The Chinese version of this paper is published in Acta Horticulturae Sinica. doi:10.16420/j.issn.0513-353x.2015-0301 http://dx.doi.org/10.1016/j.hpj.2016.07.004 2468-0141/© 2016 Chinese Society for Horticultural Science (CSHS) and Institute of Vegetables and Flowers (IVF), Chinese Academy of Agricultural Sciences (CAAS). This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/). Table 1 Chromosome parameters of 4 G. rigens varieties Chromosome No. Hongwen Xingbai Richu Zhongguo Xunzhangju Relative length/% Arm Type ratio 1 12.24 1.34 m 11.51 1.98 sm 11.40 1.33 m 5.86 1.03 m 2 11.74 1.07 m 11.06 2.14 sm 11.29 1.05 m 5.79 1.06 m 3 10.74 1.19 m 11.48 1.28 m 11.00 1.21 m 5.69 1.09 m 4 10.55 1.42 m 11.41 1.18 m 10.50 1.11 m 5.61 1.27 m 5 10.54 1.27 m 10.30 1.36 m 10.04 1.15 m 5.61 1.25 m 6 9.67 1.39 m 10.01 1.37 m 9.90 1.27 m 5.46 1.07 m 7 9.55 2.05 sm 9.77 1.34 m 9.53 1.11 m 5.43 1.05 m 8 9.51 2.68 sm 9.06 1.17 m 9.17 1.18 m 5.38 1.08 m 9 8.27 1.53 m 7.91 1.17 m 9.01 1.21 m 5.33 1.62 m 10 7.19 1.15 m 7.47 1.57 m 8.15 1.04 m 5.23 1.65 m 11 5.10 1.44 m 12 5.01 1.16 m 13 5.02 2.40 sm 14 4.21 2.62 sm 15 4.74 1.02 m 16 4.62 1.19 m 17 4.50 1.15 m 18 4.16 1.32 m 19 3.77 1.07 m 20 3.49 1.13 m Table 2 Karyotype characteristics of 4 G. rigens v arieties Variety Chromosome number Type Relative length/% Arm ratio Type Relative length/% Arm ratio Type Relative length/% Arm ratio acid for 8–10 min using a Thermostatic Water Bath at 60 °C, removed and washed three times with pure water (with each washing lasting 1–2 min). Roots were then dyed with aceto- carmine for 5–10 min, and 1–2 mm slices of root tip were placed on microscope slides and covered with coverslips. The bubble inside was extruded by tapping slightly. It was then dried with an alcohol lamp 1–2 times, and then observed and photo- graphed using a Leica DM300 microscope. About 40–50 clearly viewable cells of each variety were ob- served at the metaphase. We selected cells whose number of consistent chromosomes exceeded 85%. We photographed root tip cells, at mitosis metaphase, that had well-separated chromo- somes and a clear centromere. Chromosomes were measured and matched using Photoshop CS 5.0 software and their relative length, arm ratio, chromosome length ratio were calculated (Zhou et al., 2009; Lu et al., 2013). The Li and Chen (1985) method was used for karyotype analysis and the Stebbins (1971) method was used for karyotype classification. The karyotype asymme- try coefficient (As. K, %) was defined as (length of long arm/ length of the entire chromosome) × 100 (Arano, 1963). 3. Results Metaphase karyotype analysis of root tip cells showed that centromeres were mainly located at the middle (m) or near the Centromere location Chromosome Asymmetrical Percent with Arm Karyotype m sm length ratio (longest/shortest) karyotype arm ratio >2 index classification coefficient/% Hongwen 10 8 2 1.70 58.80 20.00 20 2A Xingbai 10 8 2 1.54 58.84 10.00 20 2A Richu 10 10 1.40 53.80 0 20 1A Zhongguo Xunzhangju 20 18 2 1.68 55.86 10.00 40 2A 280 ZENG Jiashi et al. middle (sm) of chromosomes. There was little variation of chro- mosome length. The number of chromosomes with length ratios of 1.40–1.70, and arm ratios >2:1, accounted for 0–20.00% of the total and their asymmetrical karyotype coefficients were 53.80%–58.84%. In the karyotypes of the four G. rigens vari- eties, the chromosomes are arranged in descending order (Fig. 1, Table 1 and Table 2). The chromosome number of ‘Hongwen’ was 2n = 10. The karyotype formula was 2n = 8m + 2sm, of which the fourth pair of chromosomes was the sm type (mean arm ratio = 2.34) (Fig. 1, Table 1). The ratio of the longest and shortest chromosome was 1.70, frequency of chromosomes with arm ratio >2 was 20.00%, and the karyotype was ‘2A’ (Table 2). The chromosome number of ‘Xingbai’ was 2n = 10. The karyo- type formula was 2n = 8m + 2sm. The first pairs of chromosomes were type sm (mean arm ratio = 2.06) (Fig. 1, Table 1). The asym- metrical karyotype coefficient was the maximum (58.84%) and the karyotype was ‘2A’ (Table 2). The chromosome number of ‘Richu’ was 2n = 10. All chro- mosomes were type m, and the karyotype formula was 2n = 10m (Fig. 1, Table 1). The ratio of the longest and shortest chromo- some, asymmetrical karyotype coefficient, and percentage of chromosomes exceeding 2:1 of the arm ratio were respectively 1.40%, 53.80% and 0. The karyotype was ‘1A’ (Table 2). as 2n = 20. The seventh pair of chromosomes was type sm mean arm ratio = 2.50), and the karyotype formula was 2n = 18m 2sm (Fig. 1, Table 1). Length ratio of the chromosome was 1.68. he number of chromosomes with more than 2:1 of arm ratio counted for 10.00% of the total, the asymmetrical karyotype efficient was 55.86%, and the karyotype was ‘2A’ (Table 2). Discussion Chen et al. (2003) suggested that the chromosome rdinal number of Gazania w as x = 10, karyotype formula was 2n = 2x = 18m + 2sm, and the karyotype was ‘2A’, which corresponded to the karyotype of ‘c’ in this study. However, our results show that somatic cells of three G. rigens v arieties, introduced from Japan, had ten chromosomes. It is possible that the chromosome cardinal number of Gazania w as x = 5, and ‘Zhongguo Xunzhangju’ is a tetraploid. The chromosome morphology differed among the varieties. The main chromosome type was m, and in addition, ‘Hongwen’, ‘Xingbai’ and ‘Zhongguo Xunzhangju’ each had a pair of sm chromosomes. Based on karyotype characters, only ‘Richu’ was a ‘1A’ type, while the other three varieties were the ‘2A’ type The chromosome number of ‘Zhongguo Xunzhangju’ 281 Karyotype Analysis of Gazania rigens V arieties Fig. 1 Metaphase chromosome maps (left), karyogram (middle) and ideogram (right) of 4 G. rigens varieties with a relatively symmetrical karyotype. The diversity of karyo- ). Li et al. (1983) and Wang et al. (1991) suggested that the types corresponds to the phenotypic variation in these G. rigens nal number of chromosomes in wild species of the genus varieties. dranthema was x = 9, which exhibited euploidy variation with We found poor germination in the filial generation of species being tetraploid and hexaploid. The cultivars of this ‘Zhongguo Xunzhangju’ and the other Japanese Gazania. This s mostly had aneuploid variation based on hexa- ploidy. Li et may be related to the sterility caused by a previous hybridiza- tion 007) researched the karyotypes of fourteen chrysanthemum ties. The different cultivars were all aneu- ploids with event or low triploid fertility. A major component of evolution involves the mosome numbers between 48 and 56 compared to hexaploid quantitative and structural variation of chromosomes (Li et al., = 54). Zhu et al. (2011) found that the vast majority of ional Chinese chrysanthemum varieties were hexaploid or aneuploid based on hexaploid, while a few variet- ies had SNG201409). aneuploid variation based on septuploidy and octoploidy. In addition, the ascending or descending order of aneuploid varia- References tion in plant chromosome cardinal numbers led to quantitative and rano, H., 1963. Cytological studies in subfamily Carduoideae (Compositae) structural variation of chromosomes, and played a signifi- cant role of Japan. IX. Bot Mag (Tokyo), 76: 32–39. Barkley, T.M., 2006. Flora in intraspecific differentiation and speciation (Li, 1988). Wang and f North America. Oxford University Press, New Li (1987) found that evolution of Sanvitalia was af- fected by York, 19: 196. Chen, R.Y., Song, W.Q., Li, X.L., Li, M.X., Liang, G.L., ascending aneuploid variation on cardinal chromosomes. The hen, C.B., 2003. Chromosome atlas of major economic plants genome in China, original cardinal number in Sanvitalia was x = 9 and there were ol. III. Science Press, Beijing, pp. 207–208. Chu, R.Y., Sun, X.X., 1986. The secondary cardinal numbers, x = 12 and x = 13. Zhang and Gu udies on cytogenetics of plants of the genus Morus: I. The number of hromosome of some mulberry varieties. Acta Sericol Sin, 12: 199–202. (in (2005) pointed out that there was descending aneu- ploid variation hinese) Guo, H.R., Yu, M.D., 2000. The study of karyotype of cell chromosome on cardinal chromosomes of Streptopus. In addition to two known f artificial triploid mulberry variety 9204. Acta Sericol Sin, 26: 65–70. (in cardinal numbers (x = 8 and x = 9), a new one, x = 7, was found. hinese) He, N.J., Zhang, C., Qi, X.W., Zhao, S.C., Tao, Y., Yang, G.J., Lee, T.H., And it seemed that the cardinal number of x = 7 was merged by Wang, X.Y., Cai, Q.L., Dong, L., Lu, M.Z., Liao, S.T., Luo, G.Q., He, R.J., Tan, the centromeres of a pair of large m-type chromosomes in genome ., Xu, Y.M., Li, T., Zhao, A.C., Jia, L., Fu, Q., Zeng, Q.W., Gao, C., Ma, B., iang, J.B., Wang, X.L., Shang, J.Z., Song, P.H., Wu, H.Y., Fan, L., Wang, Q., of x = 8. Chu and Sun (1986) consid- ered that the chromosome huai, Q., Zhu, J.J., Wei, C.J., Zhu, S.K., Jin, D.C., Wang, J.P., Tao, L., Yu, M.D., cardinal number of Morus w as x = 14, which was widely cited ang, C.M., Wang, Z.J., Dai, F.W., Chen, J.F., Liu, Y., Zhao, S.T., Lin, T.B., (Yu, 1986; Yu et al., 1996; Guo and Yu, 2000). However, He et al. hang, S.G., Wang, J.Y., Wang, J., Yang, H.M., Yang, G.W., Wang, J., Paterson, (2013), on a whole genome se- quencing study of Morus notabilis, .H., Xia, Q.Y., Ji, D.F., Xiang, Z.H., 2013. Draft genome sequence of the found that chromosome cardinal number of this genus should be x mulberry tree Morus notabilis. Nat Commun, 4: 2445. ttp://www.nature.com/ncomms/2013/130919/ncomms3445/full/ncomms3445 = 7. Li et al. (2014) showed that there were fourteen chromosomes tml. Li, C., Chen, F.D., Zhao, H.B., Chen, S.M., 2008. Karyotype diversity of 17 in the somatic cells of Morus yunnanensis. Therefore, our study hrysanthemum cultivars with small inflorescences. Acta Hortic Sin, 35: 71–80. Li, suggests that G. rigens introduced from Japan may be the original ., Chen, F.D., Zhao, H.B., Fang, W.M., 2007. Karyomorphology of fourteen type of the Chinese varieties, with relatively simple flower shape cultivars in cut chrysanthemum. Acta Hortic Sin, 34: 1235–1242. Li, M.X., 1988. Different euploidy variation and evolution of chromosome and color, and lower ploidy (2n = 2x = 10). Most Chinese varieties cardinal number on plants. Bull Biol, 7: 7–12. (in Chinese) Li, M.X., have large, colorful and complex flowers, and ploidy values of 2n hen, R.Y., 1985. A suggestion on the standardization of karyotype = 4x = 20; these existing varieties were formed by artificial analysis in plants. Plant Sci J, 3: 297–302. (in Chinese) Li, M.X., selection after natural variation and cross-pollination. hang, X.F., Chen, J.Y., 1983. Cytological studies on some Chinese wild It is not uncommon to see plant chromosome cardinal endranthema s pecies and chrysanthemum cultivars. Acta Hortic Sin, 10: numbers, ascending or descending, produced by aneuploid 99–205. Li, Y., Xuan, Y.H., Wang, S., Zeng, Q.W., Xiang, Z.H., He, N.J., 2014. variation among species within a genus. There are derivative or aryotype analysis of Morus yunnanensis. Sci Sericult, 40: 4. (in Chinese) Li, secondary car- dinal numbers of aneuploid variation based on .F., 2011. Study on introduction and application of ground-cover plant Gazania original cardinal number in the Asteraceae. Barkley (2006) gens L . in Suzhou area [M. D. Dissertation]. Soochow University, Suzhou. (in determined that the chromosome cardinal number of G. rigens was hinese) Lu, Y.F., Jiang, J.X., Yi, Z.L., 2013. The protocol for processing of the x = 9 (2n = 18), which may be generated by descending aneuploid hromosomal images of Miscanthus for karyotype analysis using Adobe Photoshop variation based on a tetraploid species. In contrast, Chen et al. oftware. Pratacult Sci, 30: 922–926. (in Chinese) Stebbins, G.L., 1971. (2003) sug- gested that the chromosome cardinal number of hromosomal evolution in higher plants. Edward Arnold, London, pp. 87–93. Wang, J.W., Yang, J., Li, M.X., 1991. Karyotype Gazania w as udy of five species of Chinese x = 10, but this might not be the original cardinal number of this Dendranthema. Acta Bot Yunnanica, 13: 411–416. (in Chinese) Wang, genus. Specification of the true Gazania chromosome cardinal W., 2013. Studies on sexual reproduction biology and progenies phenotype analysis number clearly needs further research. f Gazania rigens L . [M. D. Dissertation]. Soochow University, Suzhou. (in Acknowledgments hinese) Wang, X.L., Li, M.X., 1987. Observation of chromosome on 10 ompositae This work was financially supported by Project forspecies. Plant Sci J, 5: 111–118. (in Chinese) Suzhou Science and Technology Support Program (Agriculture) 282 ZENG Jiashi et al. Wang, Y.L., 2012. Studies of resistance physiology and landscape application of ingbai’Gazania [ M. D. Dissertation]. Soochow University, Suzhou. (in Chinese) Streptopus (Liliaceae). Acta Phytotaxon Sin, 43: 533–538. (in Chinese) Zhou, J.S., Su, X.B., Tang, Y.P., Luo, S.C., Zhan, F.X., Chen, G.Y., 2009. Analysis of chromosomal karyotype of Asparagus by use of Software Photoshop. Acta Agric Jiangxi, 21: 73–75. (in Chinese) Zhou, X.H., 2014. Studies on new cold resistant cultivars breeding and low carbon cultivation techniques of Gazania rigens L . [M. (4): 6. (in Chinese) Yu, M.D., Jing, C.J., Chen, W., Li, Q.R., Liao, D. Dissertation]. Soochow University, Suzhou. (in Chinese) Zhu, M.L., Liu, Q.Q., W.M., Dai, C.X., Zhao, H., Pan, J., Liu, M.X., Li, W., 1996. Studies on chromosome ploidy of some germplasms maintained at mulberry resources centre Dai, S.L., 2011. Karyotype analysis of 38 large-flowered chrysanthemum cultivars from China. Chin Bull Bot, 46: 447–455. (in Chinese) of Sichuan Province. Newsl Sericult Sci, 16: 2–5. (in Chinese) Xie, L.M., Hu, J.X., Huang, W.C., 2013. Discussion on cutting and rapid propagation technology of Gazania rigens L. Jiangsu Agric Sci, 41: 167–168. (in Chinese) Yu, M.D., 1986. Study on chromosome ploidy of mulberry. Newsl Sericult Sci, Zhang, T., Gu, Z.J., 2005. A new basic chromosome number of x = 7 for the genus 283 Karyotype Analysis of Gazania rigens V arieties